Abstract
Introduction
Acute myeloid leukemia (AML) is a heterogenous hematologic malignancy that primarily affects older adults, with a median age of diagnosis at 68 years and five-year relative survival of 29.5%. The incidence of AML diagnosis is expected to increase with an aging population in the United States, encouraging the exploration for treatment risk stratification to inform care. One factor impacting survivorship is treatment-related toxicity. Cytarabine (active metabolite, Ara-C) continues as a mainstay agent for initial induction regimens in AML, but efforts to minimize its toxicity remain a challenge. Interpatient variability presents another intricacy, where response to cytarabine can manifest as a subset of patients who have an inadequate response or those who are overly sensitive to its effects. Pharmacogenes in the cytarabine pathway may be responsible for this inconsistent response and could influence toxicity susceptibility.
We conducted a retrospective analysis of AML patients treated with cytarabine to evaluate whether single nucleotide polymorphisms (SNPs) in its pharmacogenes are associated with treatment-related mucositis. Specifically, mucositis can negatively impact quality of life with pain and difficulty eating and swallowing, Thus, identifying patients likely to develop mucositis can assist in timely supportive care.
Methods
Patients from the University of Florida Shand's Hospital who received cytarabine during their first induction therapy from 2009 to 2019 for de novo AML were screened for mucositis within the first thirty days. We obtained FFPE tissues of these patients and extracted genomic DNA. Using TaqMan real-time PCR assays (QuantStudio 3) in-house, we genotyped for ten SNPs from genes involved in the cytarabine metabolic pathway. Logistic regression models were used to test for association between these SNPs and the incidence of mucositis. All analyses were performed using Rstudio v4.1.0. SNPs with significant results were then tested for other SNPs that occur in linkage disequilibrium (LD) using HaploReg v4.1, in addition to testing for association with gene expression using the Genotype-Tissue Expression Portal (GTEx).
Results
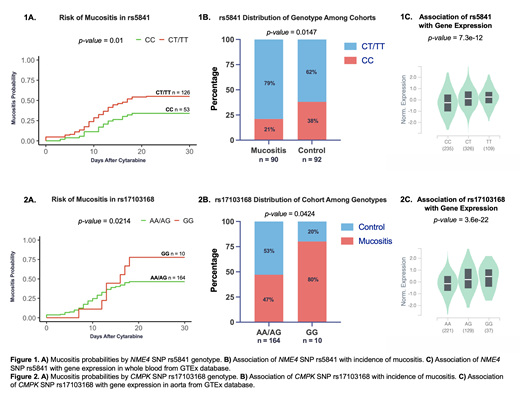
In total, 184 patients were included in this study, with 92 in each group - mucositis and no mucositis (control). The median age was 64 years, 58% were male, 83% were white, 12% African American or black, and 0.2% were Asian. No difference in the incidence of mucositis was observed by sex or race. Logistic regression models identified two SNPs significantly associated (p<0.05) with incidence of mucositis within the first thirty days of cytarabine exposure. For rs5841, a synonymous coding SNP in NME4, presence of the variant T allele (CT/TT genotype) was associated with significantly increased incidence of mucositis (OR=1.51; 95% CI [1.09-2.11]) (Figure 1A and B). rs5841 occurs in LD with seven other SNPs impacting multiple regulatory motifs. Consistent with these results, presence of the T allele is associated with higher NME4 expression (Figure 1C), implying that higher ara-C activation in the variant T group is contributing to greater incidence of mucositis. For rs17103168, an intronic SNP in CMPK, patients homozygous for the variant allele (GG genotype) experienced significantly higher incidence of mucositis compared to those with AG or AA genotypes (OR=1.56; 95% CI [0.95-2.58]) (Figure 2A and B). This SNP occurs in LD with sixty other SNPs impacting numerous regulatory motifs as well. Presence of the GG genotype is associated with high gene expression (Figure 2C), suggesting that this increased gene expression in GG genotypes may result in higher ara-C levels, and subsequently increasing risk of toxicities.
Conclusion
Overall, we report a significant association between the risk of mucositis after initial cytarabine exposure and two SNPs in cytarabine metabolic pathway genes. Identifying those who may experience adverse toxicity using such SNP-based prediction models will allow for clinically meaningful interventions. Consideration of interpatient variability with cytarabine may lend valuable insight in shared-decision making between patient and clinician when weighing risks vs. benefit of treatment. Ongoing and future studies are focused on expanding this cohort to include evaluation of cytarabine pharmacogenomics with respect to disease progression and survival outcome in AML.
No relevant conflicts of interest to declare.